Motivation
As I stated in this post yesterday, I was able to run a program using OpenMPI on a Docker container running on multiple nodes. I wanted to find out how much performance I could improve by using OpenMPI, so I decided to benchmark it. Actually, I had some difficulties this time as well, and I would be happy if that part is helpful for others.
Sources
- Himeno Benchmark This is the home page of the HIMENO Benchmark.
- Horovod with MPI This page has answers to the points I struggled with this time.
- Solution to the problem of OpenMPI(v3.1.3) not being able to parallelize across multiple nodes I think I found the same phenomenon and solution. However, this page is a solution on a physical machine.
Download, Running on a single machine
Download
Download “C+MPI, static allocate version” from the download page. lzh compressed, install the jlha-utils package and extract the downloaded lzh file.
$ sudo apt install jlha-utils
$ lha x cc_himenobmtxp_mpi.lzh
From unzipping to running on a single machine, follow the instructions in the “How to use” section of the download page.
I copied the unzipped file to /ext/nfs/athena++/himeno and worked from there. As explained yesterday, this area is NFS mounted from other nodes.
From now on, the work was done in a docker container. paramset.sh was granted execution rights.
# pwd
/workdir/himeno
# mv param.h param.h.org
# ./paramset.sh M 1 1 2
# cp Makefile.sample Makefile
# ls -l
total 44
-rw-rw-r-- 1 1000 1000 251 Feb 22 12:24 Makefile
-rw-rw-r-- 1 1000 1000 251 Feb 22 12:20 Makefile.sample
-rw-rw-r-- 1 1000 1000 4414 Feb 22 12:19 cc_himenobmtxp_mpi.lzh
-rw-rw-r-- 1 1000 1000 13099 Feb 22 12:20 himenoBMTxps.c
-rw-rw-r-- 1 root root 196 Feb 22 12:23 param.h
-rw-rw-r-- 1 1000 1000 202 Feb 22 12:20 param.h.org
-rwxrwxr-x 1 1000 1000 2079 Feb 22 12:20 paramset.sh
# make
Make will output warnings, but a.out called bmt can be created and run on a single machine.
# mpirun -np 2 ./bmt
Authorization required, but no authorization protocol specified
Authorization required, but no authorization protocol specified
Sequential version array size
mimax = 129 mjmax = 129 mkmax = 257
Parallel version array size
mimax = 129 mjmax = 129 mkmax = 131
imax = 128 jmax = 128 kmax =129
I-decomp = 1 J-decomp = 1 K-decomp =2
Start rehearsal measurement process.
Measure the performance in 3 times.
MFLOPS: 9847.792258 time(s): 0.041767 1.667103e-03
Now, start the actual measurement process.
The loop will be excuted in 4309 times
This will take about one minute.
Wait for a while
cpu : 64.531007 sec.
Loop executed for 4309 times
Gosa : 1.978532e-04
MFLOPS measured : 9155.072245
Score based on Pentium III 600MHz : 110.515116
After all this time, and with yesterday’s experience, I thought it would be easy to run BENCHMARK on multiple nodes.
More Processes Running Concurrently
Now that I was able to run two processes on a single machine, I decided to run four processes. To do so, I ran the following. For the meaning of the parameters, please refer to the download page.
# cat hosts.txt
europe
ganymede
jupiter
# ./paramset.sh M 1 2 2
# make
# mpirun --hostfile hosts.txt -mca plm_rsh_args "-p 12345" -np 4 $(pwd)/bmt
The results are as follows
Authorization required, but no authorization protocol specified
Authorization required, but no authorization protocol specified
Authorization required, but no authorization protocol specified
Authorization required, but no authorization protocol specified
Authorization required, but no authorization protocol specified
Authorization required, but no authorization protocol specified
Invalid number of PE
Please check partitioning pattern or number of PE
To find out what the PE is at this time (expected value is 1x2x2=4), I added the following to the part that is giving the error.
printf("PE=%d\n",npe);
Then I noticed when I made it! Even though param.h was updated, himenoBMTxps.c was not compiled (himenoBMTxps.o was not updated) and bmt was created. The following will work.
# ./paramset.sh M 1 2 2
# make clean
# make
# mpirun --hostfile hosts.txt -mca plm_rsh_args "-p 12345" -np 4 $(pwd)/bmt
Running on multiple nodes
I decided to run the BENCHMARK by varying the number of processes to 2, 4, 8, 16, 32, and 64. The computation size is set to L.
First, the number of processes was set to 8, which is the middle number of processes. The file “. /maramset.sh L 2 2 2 2” to update param.h, and then “make clean” and “make” were executed. Also, myhosts at this time is the same as the hosts.txt file mentioned above.
# mpirun -np 8 --hostfile myhosts -mca plm_rsh_args "-p 12345" -oversubscribe $(pwd)/bmt
Authorization required, but no authorization protocol specified
Authorization required, but no authorization protocol specified
Authorization required, but no authorization protocol specified
Authorization required, but no authorization protocol specified
Authorization required, but no authorization protocol specified
Authorization required, but no authorization protocol specified
[ganymede][[22394,1],4][../../../../../../opal/mca/btl/tcp/btl_tcp_endpoint.c:625:mca_btl_tcp_endpoint_recv_connect_ack] received unexpected process identifier [[22394,1],7]
I searched for “received unexpected process identifier” and found a hit on the page of source 3.
After reading the solution “Add --mca btl_tcp_if_include eth0 to the runtime options” on this page, I remembered Source 2 and was able to solve the problem as follows.
# mpirun -np 8 --hostfile myhosts -mca plm_rsh_args "-p 12345" -mca btl_tcp_if_exclude lo,docker0 $(pwd)/bmt
Authorization required, but no authorization protocol specified
Authorization required, but no authorization protocol specified
Authorization required, but no authorization protocol specified
Authorization required, but no authorization protocol specified
Authorization required, but no authorization protocol specified
Authorization required, but no authorization protocol specified
Sequential version array size
mimax = 257 mjmax = 257 mkmax = 513
Parallel version array size
mimax = 131 mjmax = 131 mkmax = 259
imax = 129 jmax = 129 kmax =257
I-decomp = 2 J-decomp = 2 K-decomp =2
Start rehearsal measurement process.
Measure the performance in 3 times.
MFLOPS: 19912.780454 time(s): 0.168541 8.841949e-04
Now, start the actual measurement process.
The loop will be excuted in 1067 times
This will take about one minute.
Wait for a while
(Omitted below)
Again, the Horovod in Docker experience was helpful.
Performance measurements
The following measurements were taken on five machines. The hostfile at this time was as follows.
# cat myhosts
europe
jupiter
ganymede
saisei
mokusei
No slot specified
| np | MFLOPS |
|---|---|
| 2 | 9,177 |
| 4 | 11,267 |
| 8 | 20,501 |
| 16 | 20,111 |
| 32 | 20,607 |
| 64 | 8,411 |
I thought it might be strange that they are saturated at np=8. As you can see from the table below (an excerpt of the specs for each machine), the total number of physical cores (# of CPU x # of cores) for the 5 machines is 26, so I thought 16 would be the peak to saturate.
| jupiter | ganymede | saisei | mokusei | europe | |
|---|---|---|---|---|---|
| CPU | Xeon(R) CPU E5-1620 @ 3.60GHz | Xeon(R) CPU E5-2620 @ 2.00GHz | Xeon(R) CPU E5-2643 @ 3.30GHz | Xeon(R) CPU E5-2609 @ 2.40GHz | eon(R) CPU E3-1270 v5 @ 3.60GHz |
| # of CPU | 1 | 1 | 1 | 2 | 1 |
| # of core | 4 | 6 | 4 | 4 | 4 |
with slot specified
I decided to list the number of physical cores of each machine as slots in the hostfile.
# cat myhosts
europe slots=4
jupiter slots=4
ganymede slots=6
saisei slots=4
mokusei slots=8
The results are as follows. The results are in line with the expected trend.
| np | MFLOPS |
|---|---|
| 2 | 9,171 |
| 4 | 11,234 |
| 8 | 20,413 |
| 16 | 29,589 |
| 32 | 23,018 |
| 64 | 9,216 |
The measurement results are graphed as follows.
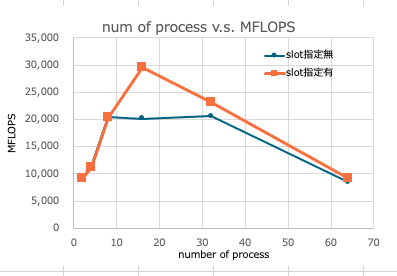
Summary
I thought that the performance would improve as the number of parallel processes increased up to the number of physical cores, and I obtained such results. I do not know the reason for the difference in the peak number of processes depending on whether the slot specification is used or not.
I measured the calculation size as “L” this time, but I would like to investigate how the difference in calculation size affects the results in the future.