Introduction
Up to this article posted last month, I have confirmed that OpenMPI can be embedded in a Docker container and used for parallel computing on multiple nodes. In this post, I will use the Docker container created above to run tutorial 4 “Running 3D MHD with OpenMP and MPI” of Athena++ on multiple nodes.
Sources.
- Athena Tutorial This time, I will run “4. Running 3D MHD with OpenMP and MPI”.
- Japanese page of the above This page is maintained by Dr. Kengo Tomida.
- H5Pset_fapl_mpio was not declared in this scope A page I searched to investigate the cause of the compile error and found that the parallel HDF5 library is required.
- h5fortran-mpi A page that I found by searching for “parallel HDF5 library”. In ubuntu, that is libhdf5-mpi-dev package.
Execution Environment
Dockerfile
The Dockerfile to run this tutorial is as follows
# Based on the Dockefile for creating a Docker image that JupyterLab can use,
# Create a Docker container that can be used for athena++ development and execution.
# Based on the latest version of ubuntu 22.04.
FROM ubuntu:jammy-20240111
# Set bash as the default shell
ENV SHELL=/bin/bash
# Build with some basic utilities
RUN apt update && apt install -y \
build-essential \
python3-pip apt-utils vim \
git git-lfs \
curl unzip wget gnuplot \
openmpi-bin libopenmpi-dev \
openssh-client openssh-server \
libhdf5-dev libhdf5-openmpi-dev
# alias python='python3'
RUN ln -s /usr/bin/python3 /usr/bin/python
# install python package to need
RUN pip install -U pip setuptools \
&& pip install numpy scipy h5py mpmath
# The following stuff is derived for horovod in docker.
# Allow OpenSSH to talk to containers without asking for confirmation
RUN mkdir -p /var/run/sshd
RUN cat /etc/ssh/ssh_config | grep -v StrictHostKeyChecking > /etc/ssh/ssh_config.new && \
echo " StrictHostKeyChecking no" >> /etc/ssh/ssh_config.new && \
mv /etc/ssh/ssh_config.new /etc/ssh/ssh_config
# --allow-run-as-root
ENV OMPI_ALLOW_RUN_AS_ROOT=1
ENV OMPI_ALLOW_RUN_AS_ROOT_CONFIRM=1
# Set hdf5 path
ENV CPATH="/usr/include/hdf5/openmpi/"
# Create a working directory
WORKDIR /workdir
# command prompt
CMD ["/bin/bash"]
In the end, using the container created from the above Dockerfile, I was able to run this tutorial. The trial and error process to obtain this Dockerfile is described below. The point was to be able to use HDF5 in an MPI environment.
A record of the errors I encountered before creating a container to use HDF5.
The first step is to make the following configuration.
# python configure.py --prob blast -b --flux hlld -mpi -hdf5
Your Athena++ distribution has now been configured with the following options:
Problem generator: blast
Coordinate system: cartesian
Equation of state: adiabatic
Riemann solver: hlld
Magnetic fields: ON
Number of scalars: 0
Number of chemical species: 0
Special relativity: OFF
General relativity: OFF
Radiative Transfer: OFF
Implicit Radiation: OFF
Cosmic Ray Transport: OFF
Frame transformations: OFF
Self-Gravity: OFF
Super-Time-Stepping: OFF
Chemistry: OFF
KIDA rates: OFF
ChemRadiation: OFF
chem_ode_solver: OFF
Debug flags: OFF
Code coverage flags: OFF
Linker flags: -lhdf5
Floating-point precision: double
Number of ghost cells: 2
MPI parallelism: ON
OpenMP parallelism: OFF
FFT: OFF
HDF5 output: ON
HDF5 precision: single
Compiler: g++
Compilation command: mpicxx -O3 -std=c++11
# make clean
rm -rf obj/*
rm -rf bin/athena
rm -rf *.gcov
hdf5.h: No such file or directory
# make
mpicxx -O3 -std=c++11 -c src/globals.cpp -o obj/globals.o
mpicxx -O3 -std=c++11 -c src/main.cpp -o obj/main.o
In file included from src/main.cpp:46:
src/outputs/outputs.hpp:22:10: fatal error: hdf5.h: No such file or directory
22 | #include <hdf5.h>
| ^~~~~~~~
compilation terminated.
make: *** [Makefile:119: obj/main.o] Error 1
To address the above error, I added libhdf5-dev to the Dockerfile to install. However, this alone did not solve the error, and export CPATH="/usr/include/hdf5/serial/" was added to the Dockerfile.
After that, I made again.
#ifdef MPI_PARALLEL
# make
mpicxx -O3 -std=c++11 -c src/globals.cpp -o obj/globals.o
・・・
・・・
mpicxx -O3 -std=c++11 -c src/inputs/hdf5_reader.cpp -o obj/hdf5_reader.o
src/inputs/hdf5_reader.cpp: In function 'void HDF5ReadRealArray(const char*, const char*, int, const int*, const int*, int, const int*, const int*, AthenaArray<double>&, bool, bool)':
src/inputs/hdf5_reader.cpp:94:7: error: 'H5Pset_fapl_mpio' was not declared in this scope; did you mean 'H5Pset_fapl_stdio'?
94 | H5Pset_fapl_mpio(property_list_file, MPI_COMM_WORLD, MPI_INFO_NULL);
| ^~~~~~~~~~~~~~~~
| H5Pset_fapl_stdio
src/inputs/hdf5_reader.cpp:109:7: error: 'H5Pset_dxpl_mpio' was not declared in this scope; did you mean 'H5Pset_fapl_stdio'?
109 | H5Pset_dxpl_mpio(property_list_transfer, H5FD_MPIO_COLLECTIVE);
| ^~~~~~~~~~~~~~~~
| H5Pset_fapl_stdio
make: *** [Makefile:119: obj/hdf5_reader.o] Error 1
Since the above error was in the #ifdef MPI_PARALLEL section of the source code, I investigated whether hdf5-related modules might be required for parallel computing with OpenMPI.
I found the information in the sources 3 and 4, and installed libhdf5-openmpi-dev in the Dockerfile. I also changed the CPATH setting I added above to CPATH="/usr/include/hdf5/openmpi".
link error
When I tried to make again, I got the following error at the end of compiling and linking.
/usr/bin/ld: cannot find -lhdf5: No such file or directory
collect2: error: ld returned 1 exit status
make: *** [Makefile:114: bin/athena] Error 1
After checking under /usr/lib/x86_64-linux-gnu, where the library is stored, I guessed that the library I needed was hdf5_openmpi, and changed “-lhdf5” in the Makefile generated as a result of the configuration to “-lhdf5_openmpi”.
After above trial and error, I obtained the Dockerfile shown at the beginning of this section.
Running the simulation
Edit parameter files
As in the previous tutorial, copy the parameter file (input file) and executable to the working directory “t4” in the container. The parameter file was copied from athena/inputs/mhd/athinput.blast.
# pwd
/workdir/kenji/t4
# ls -l
-rwxr-xr-x 1 root root 3613256 Mar 2 04:11 athena
-rw-r--r-- 1 root root 2193 Mar 2 08:12 athinput.blast
This time, since the time is measured in parallel processing, I decided to set the mesh to double so that it would take a little longer. The changes to athinput.blast are as follows
10c10
< file_type = hdf5 # HDF5 data dump
---
> file_type = vtk # VTK data dump
13d12
< ghost_zones = true # enables ghost zone output
24c23
< nx1 = 128 # Number of zones in X1-direction
---
> nx1 = 64 # Number of zones in X1-direction
30c29
< nx2 = 128 # Number of zones in X2-direction
---
> nx2 = 64 # Number of zones in X2-direction
36c35
< nx3 = 128 # Number of zones in X3-direction
---
> nx3 = 64 # Number of zones in X3-direction
42,47c41
< #num_threads = 1 # Number of OpenMP threads per process
<
< <meshblock>
< nx1 = 32 # Number of zones per MeshBlock in X1-direction
< nx2 = 32 # Number of zones per MeshBlock in X2-direction
< nx3 = 32 # Number of zones per MeshBlock in X3-direction
---
> num_threads = 1 # Number of OpenMP threads per process
Measuring simulation time
Create the following shell command, run 2, 4, and 8 for the number of processes (np), and compare the cpu time recorded in log files.
# cat mpi_run
mpirun -np $1 --hostfile myhosts \
-mca plm_rsh_args "-p 12345" \
-mca btl_tcp_if_exclude lo,docker0 \
-oversubscribe $(pwd)/athena \
-i $2 > log
# cat myhosts
europe slots=4
jupiter slots=4
ganymede slots=6
# mpi_run 2 athinput.blast
# mpi_run 4 athinput.blast
# mpi_run 8 athinput.blast
Measurement results
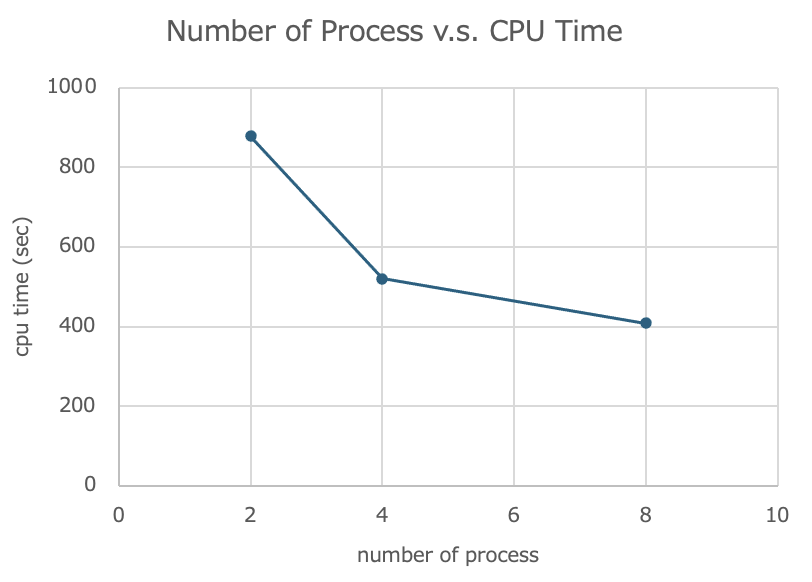
The cpu time recorded in the log was as follows.
| number of processes (np) | cpu time (sec) |
|---|---|
| 2 | 878 |
| 4 | 520 |
| 8 | 408 |
The graph is as follows.
For the future
The measurement results showed that when 8 processes are processed simultaneously, the simulation can be run in about half the time of 2 processes.
I will continue to work on the visualization part of the simulation results for this tutorial.
In addition, Dr. Tomida’s page in the source 2. has an additional task to try the simulation of “Rayleigh-Taylor instability”, which I would like to run.